
- Osirix lite nifti to dicom full version#
- Osirix lite nifti to dicom windows 10#
- Osirix lite nifti to dicom pro#
Osirix lite nifti to dicom pro#
However, if you need to adjust the export format, you just need to find the Format in the preset browser or click on the format link to reopen the Premiere Pro format dialog.

Osirix lite nifti to dicom full version#
If you are interested in this software, let’s check it out Adobe Media Encoder 2021 Free Download Full Version at the link below!Īdobe Media Encoder 2021 PC System Requirements In the latest update, Adobe claimed this software’s performance has increased up to two times faster than the previous version, and several features enhancement has been added.
Osirix lite nifti to dicom windows 10#
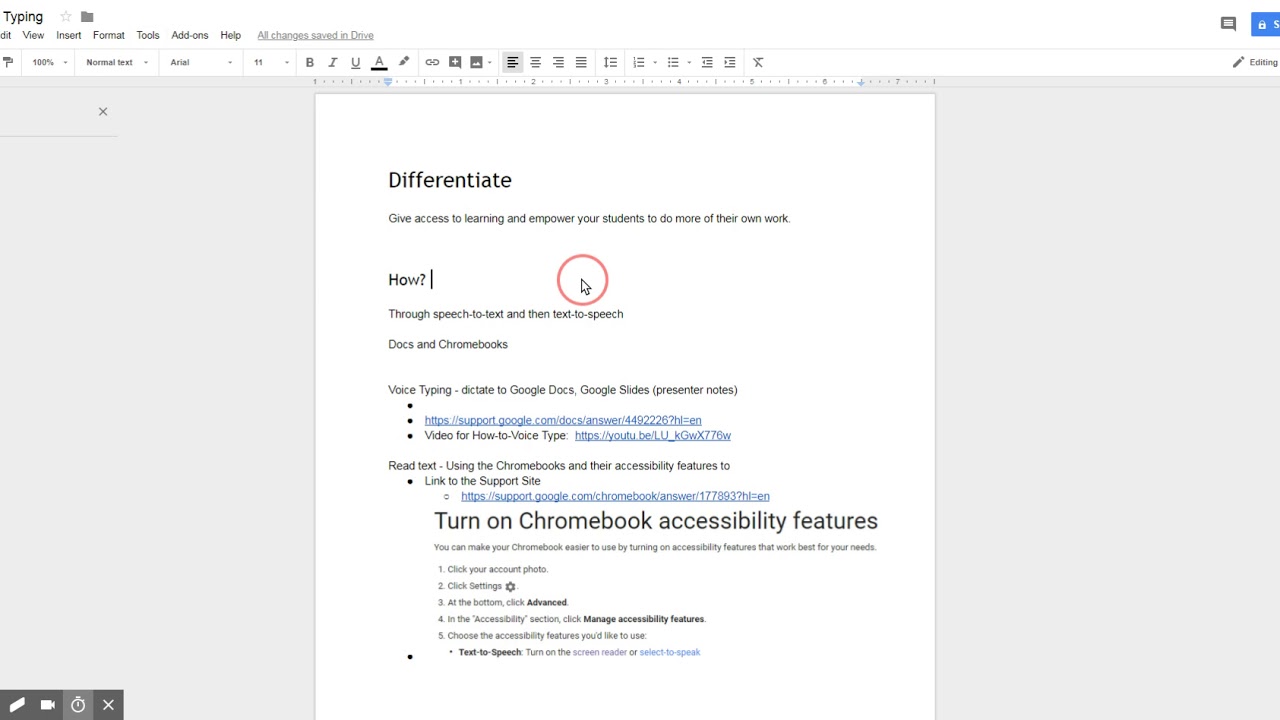
I am using this page as a source of information: Neuroimaging in Python - NiBabel 3.2.1+100.g9fdf5e3e documentationįrom what I understand, the tag 0020, 0037 provides the orientation of the rows and column of the slice of the MRI (in this case) image to transform the image into the patient (dicom) coordinate system. In my images, I get the following info from this tag:įrom my readings, I understand that the patient/world coordinate for Dicom standard is left +, posterior +, superior +. This means that the images coordinate system is left+, inferior+ and posterior+. Mask_value = mimics_mask.get_voxel_buffer() I obtain the mask using the scripting commands: Nifti patient = Dicom image where the resulting rotation matrix is: To obtain the nifti patient orientation (world), I would do the following: Therefore, i and j are inverted with respect to Dicom and the Dicom patient to Nifti patient transformation matrix is Therefore, the transformation affine matrix is from Dicom to Patient (assuming pixel spacing and slice thickness of 1 to keep it simple) and switching/flipping rows and columns as mentioned in the web page referenced above:įrom what I have read, the Nifti format has a different patient coordinate system than the Dicom, where right is +, anterior is +, and superior is+. the axial plane image (second column, bottom row) is in the locatin for the coronal image and it needs to be rotated 90 degrees the coronal plane image (second column, top row) is in the location for the sagittal image and it needs to be rotated clockwise by 90 degrees the sagittal plane image (first column, top row of the image quadrant) is in the location for the axial image However, the resulting images (showing here the mask of a deformed vertebral column) are oriented all wrong, when I view it in ITK-snap. Unfortunately we don’t have much experience with the NiBabel package and required affine transformation ourselves so I’m afraid we might be very limited in how we can help as we want to avoid giving wrong information. However, I can confirm that there is a difference between the DICOM tags and resulting Mimics project.


 0 kommentar(er)
0 kommentar(er)
